
引言
免疫细胞与脂肪细胞及其他基质成分之间相互作用,以维持脂肪组织的稳态【1】。其中,脂肪组织巨噬细胞(adipose tissue macrophages,ATMs)在调节代谢性炎症中发挥着重要作用【2-6】。巨噬细胞在不同组织中具有极强的异质性和可塑性【7,8】,而目前关于脂肪巨噬细胞的功能/表型分类过于简化。因此,鉴定在肥胖中的致病性ATMs亚群对开发靶向巨噬细胞的免疫疗法至关重要。
2024年09月17日,华中科技大学同济医学院附属同济医院王从义教授团队在Cell Metabolism发表题为PDIA3 defines a novel subset of adipose macrophages to exacerbate the development of obesity and metabolic disorders的研究论文,就PDIA3对脂肪巨噬细胞的功能调控及其在肥胖发生发展中的作用进行了深入研究。
 本研究通过对正常和肥胖患者脂肪组织样本进行单细胞测序,观察到在肥胖个体中累积了一群独特的ATMs亚群,定义为ATF4hi PDIA3hi ACSL4hi CCL2hi炎症和代谢激活的巨噬细胞(inflammatory and metabolically-activated macrophages,iMAMs)。ATF4、ACSL4和CCL2在调节巨噬细胞功能方面发挥重要作用【9-13】,而PDIA3在巨噬细胞中的作用尚未明确。PDIA3属于蛋白质二硫异构酶(protein disulfide isomerase,PDI)家族,具有异构酶和氧化还原活性,参与未折叠蛋白反应【14-16】。然而,PDIA3在肥胖及其发病机制中的作用仍未可知。
本研究通过对正常和肥胖患者脂肪组织样本进行单细胞测序,观察到在肥胖个体中累积了一群独特的ATMs亚群,定义为ATF4hi PDIA3hi ACSL4hi CCL2hi炎症和代谢激活的巨噬细胞(inflammatory and metabolically-activated macrophages,iMAMs)。ATF4、ACSL4和CCL2在调节巨噬细胞功能方面发挥重要作用【9-13】,而PDIA3在巨噬细胞中的作用尚未明确。PDIA3属于蛋白质二硫异构酶(protein disulfide isomerase,PDI)家族,具有异构酶和氧化还原活性,参与未折叠蛋白反应【14-16】。然而,PDIA3在肥胖及其发病机制中的作用仍未可知。研究团队探讨了PDIA3作为人肥胖生物标志物的可行性,并通过DIO模型、整体代谢指标监测、骨髓移植、单核细胞过继、多重转录组测序、蛋白质谱、染色质沉淀、报告基因及定点突变等实验,发现其作为效应分子调节iMAMs功能,从而加剧高脂饮食诱导的肥胖。机制层面,研究人员揭示PDIA3主要影响巨噬细胞的活化和趋化活性,巨噬细胞中敲除PDIA3可保护小鼠在高脂饮食诱导下的肥胖和相关代谢紊乱症状。具体而言,在代谢应激条件下,ATF4作为主转录因子促进PDIA3的表达。PDIA3调节RhoA活性,并通过Hippo-YAP信号通路促进巨噬细胞的活化。该研究利用纳米材料结合siRNA靶向巨噬细胞,发现PDIA3作为肥胖免疫干预靶点,具有广泛的临床转化潜力。
综上所述,本研究揭示了在肥胖个体中的一类新型的ATMs亚群,即ATF4hi PDIA3hi ACSL4hi CCL2hi炎症和代谢激活巨噬细胞(iMAMs)在肥胖中的作用及其相关机制,系统阐述了效应分子PDIA3在肥胖及其发病机制中的关键作用。研究证明分泌型PDIA3是反映肥胖代谢性炎症程度的可靠标志物,靶向抑制PDIA3有望成为治疗肥胖等代谢性疾病的新靶点。
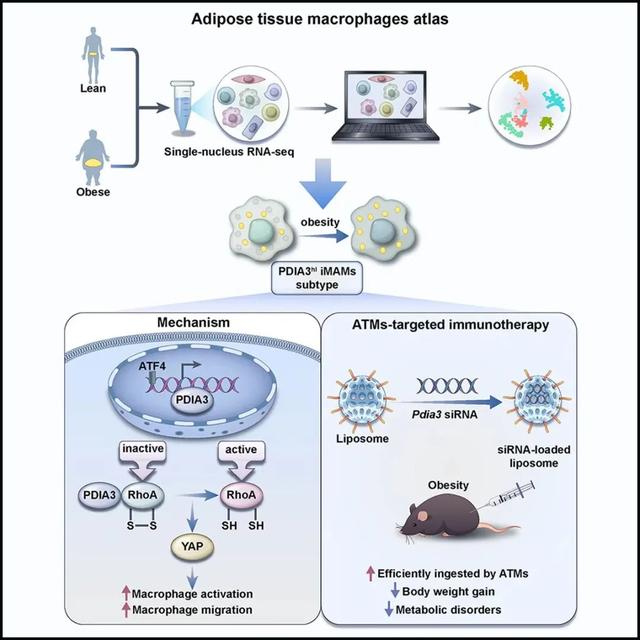
模式图(Credit: Cell Metabolism)
参考文献
1. Brestoff, J.R., and Artis, D. (2015). Immune regulation of metabolic homeostasis in health and disease. Cell 161, 146-160. 10.1016/j.cell.2015.02.022.2. Chawla, A., Nguyen, K.D., and Goh, Y.P. (2011). Macrophage-mediated inflammation in metabolic disease. Nature reviews. Immunology 11, 738-749. 10.1038/nri3071.3. Osborn, O., and Olefsky, J.M. (2012). The cellular and signaling networks linking the immune system and metabolism in disease. Nature medicine 18, 363-374. 10.1038/nm.2627.4. Odegaard, J.I., and Chawla, A. (2013). The immune system as a sensor of the metabolic state. Immunity 38, 644-654. 10.1016/j.immuni.2013.04.001.5. McNelis, J.C., and Olefsky, J.M. (2014). Macrophages, immunity, and metabolic disease. Immunity 41, 36-48. 10.1016/j.immuni.2014.05.010.6. Biswas, S.K., and Mantovani, A. (2012). Orchestration of metabolism by macrophages. Cell metabolism 15, 432-437. 10.1016/j.cmet.2011.11.013.7. Colin, S., Chinetti-Gbaguidi, G., and Staels, B. (2014). Macrophage phenotypes in atherosclerosis. Immunological reviews 262, 153-166. 10.1111/imr.12218.8. Blériot, C., Chakarov, S., and Ginhoux, F. (2020). Determinants of Resident Tissue Macrophage Identity and Function. Immunity 52, 957-970. 10.1016/j.immuni.2020.05.014.9. Iwasaki, Y., Suganami, T., Hachiya, R., Shirakawa, I., Kim-Saijo, M., Tanaka, M., Hamaguchi, M., Takai-Igarashi, T., Nakai, M., Miyamoto, Y., and Ogawa, Y. (2014). Activating transcription factor 4 links metabolic stress to interleukin-6 expression in macrophages. Diabetes 63, 152-161. 10.2337/db13-0757.10. Chen, P., Wang, D., Xiao, T., Gu, W., Yang, H., Yang, M., and Wang, H. (2023). ACSL4 promotes ferroptosis and M1 macrophage polarization to regulate the tumorigenesis of nasopharyngeal carcinoma. International immunopharmacology 122, 110629. 10.1016/j.intimp.2023.110629.11. Kuwata, H., Nakatani, E., Shimbara-Matsubayashi, S., Ishikawa, F., Shibanuma, M., Sasaki, Y., Yoda, E., Nakatani, Y., and Hara, S. (2019). Long-chain acyl-CoA synthetase 4 participates in the formation of highly unsaturated fatty acid-containing phospholipids in murine macrophages. Biochimica et biophysica acta. Molecular and cell biology of lipids 1864, 1606-1618. 10.1016/j.bbalip.2019.07.013.12. Kanda, H., Tateya, S., Tamori, Y., Kotani, K., Hiasa, K., Kitazawa, R., Kitazawa, S., Miyachi, H., Maeda, S., Egashira, K., and Kasuga, M. (2006). MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. The Journal of clinical investigation 116, 1494-1505. 10.1172/jci26498.13. Kamei, N., Tobe, K., Suzuki, R., Ohsugi, M., Watanabe, T., Kubota, N., Ohtsuka-Kowatari, N., Kumagai, K., Sakamoto, K., Kobayashi, M., et al. (2006). Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. The Journal of biological chemistry 281, 26602-26614. 10.1074/jbc.M601284200.14. Freedman, R.B., Hirst, T.R., and Tuite, M.F. (1994). Protein disulphide isomerase: building bridges in protein folding. Trends in biochemical sciences 19, 331-336. 10.1016/0968-0004(94)90072-8.15. Ellgaard, L., and Ruddock, L.W. (2005). The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO reports 6, 28-32. 10.1038/sj.embor.7400311.16. Galligan, J.J., and Petersen, D.R. (2012). The human protein disulfide isomerase gene family. Human genomics 6, 6. 10.1186/1479-7364-6-6.https://doi.org/10.1016/j.cmet.2024.08.009责编|探索君
排版|探索君
文章来源|“BioArt”
End
